Gpml2svg, a command-line/Python API for rendering GPML (GenMAPP Pathway Markup Language) to SVG, has been released today via github and PyPi.
Background
In order to add support for GPML pathways (and later KEGG) in MetaPath I needed a way to get SVG rendered versions of marked up pathways. The SVG available on WikiPathways is good but doesn't have support for external links (XRef), node identifiers/CSS (to apply transition/style effects to particular nodes). I also ideally wanted a solution that didn't rely on an internet connection to get at, and which would allow users to import GPML files directly. I saw there was a PHP-based tool for converting GPML to SVG but that it was incomplete. So I set about seeing if I could write a command-line renderer in Python: and gpml2svg is the result of doing exactly that.
Status
'Quite functional'. The two images below show (1) stock SVG available on WikiPathways
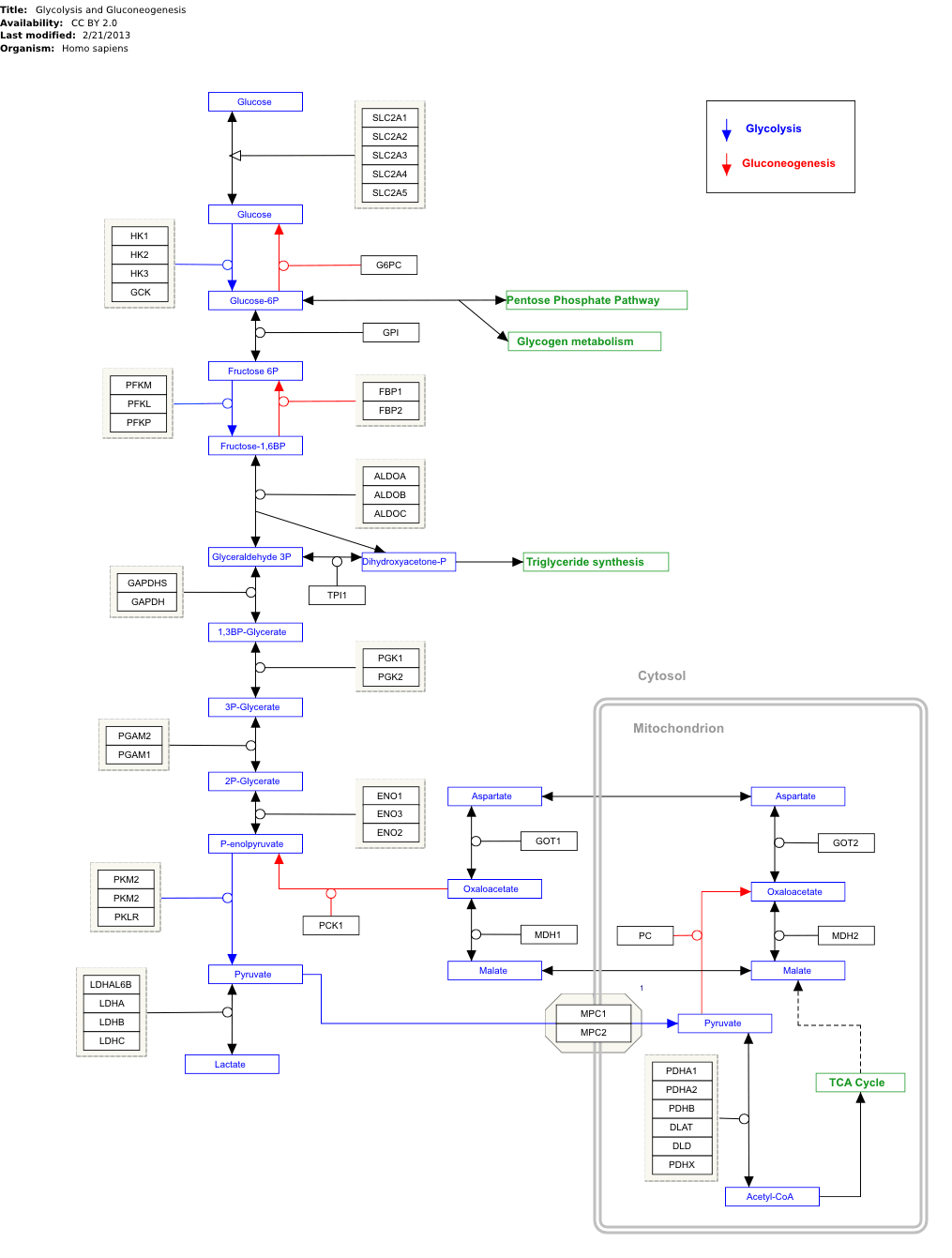
(2) output from a given GPML file using gpml2svg
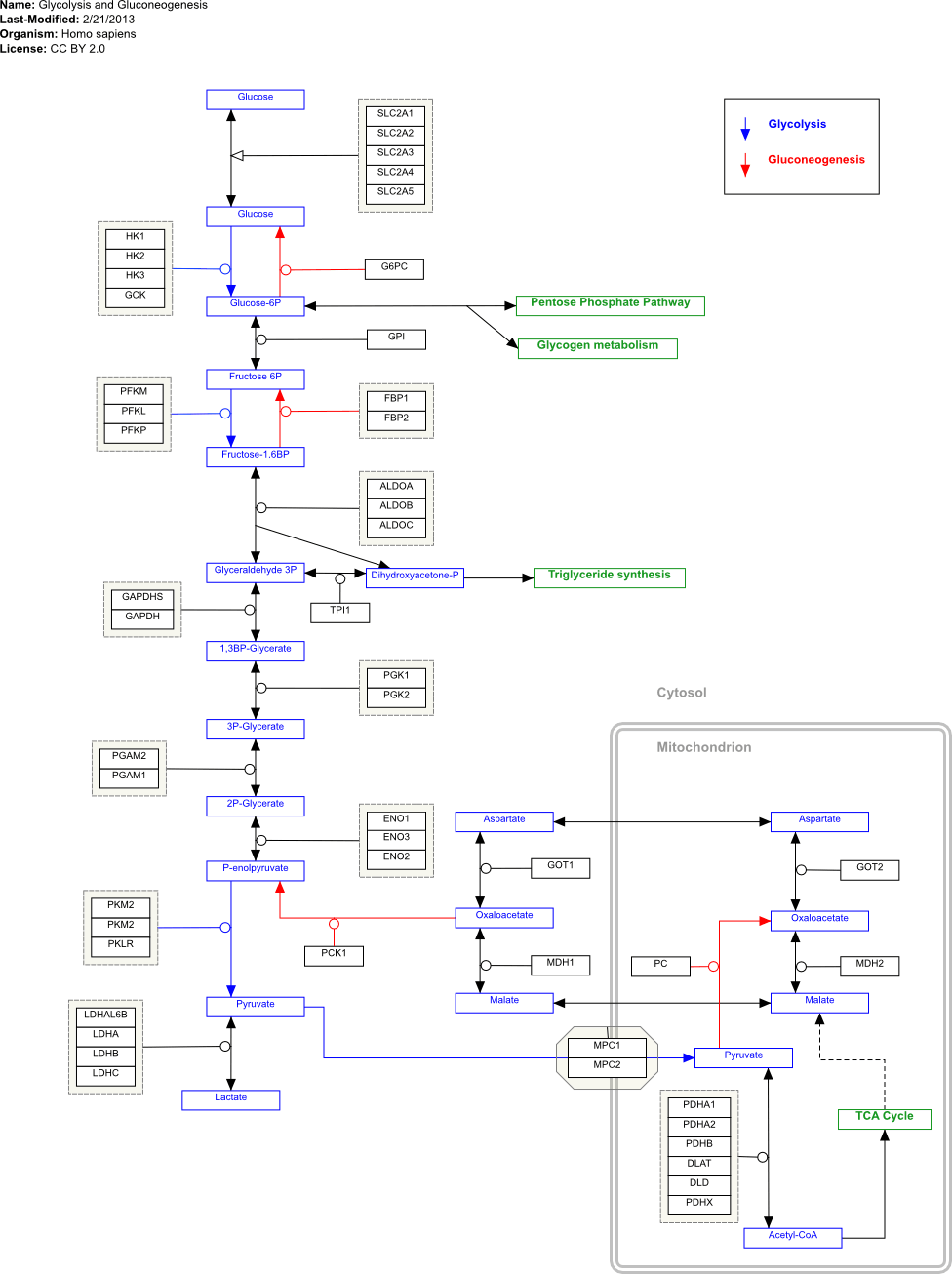
There are a few things that are 'off' (for example the mitochondrion compartment is aligned wrongly) but these are cosmetic and just need some number tweaking. There is only support for shapes that were in the files I tested (mostly squares and the group 'hexagon' for complexes). The elbow-edge drawing is also potentially awful - out of the week it took to write this I probably spent at least a third of the time on trying to get my head around making that work from the limited data in the GPML files! But it seems to be there. Hardly important but the files are also smaller.
To support developers in [[ countryRegion ]] I give a [[ localizedDiscount[couponCode] ]]% discount on all books and courses.
[[ activeDiscount.description ]] I'm giving a [[ activeDiscount.discount ]]% discount on all books and courses.
Download & Installation
You can install gpml2svg via the PyPi Python package system. You can install via the command line with:
pip install gpml2svg
Alternatively the code (and issue tracker) is available on github.
Once installed, using is as simple as entering on the command-line:
gpml2svg -f <filename.gpml>
..the resulting SVG file will be saved to the current folder with an .svg extension. The command-line interface will be improved in future!
You can also access gpml2svg as a Python package, using
from gpml2svg import gpml2svg
svg,metadata = gpml2svg.gpml2svg(<svg-as-string>)
You'll get back the rendered pathway map in svg and a dict containing name, species, revision, etc. in metadata. Other options, including color-coding and XRef extras are available - I'll be documenting these once the API has stabilised.
If you're interested in creating and editing pathways then PathVisio & WikiPathways are both great tools well worth taking a look at.
License
Gpml2svg is licensed under a Modified BSD 2 clause (i.e. completely free), my license of choice for interoperability software. You can do with it what you will, but don't come crying to me if you end up getting interested in metabolomics.
Contributions
Any suggestions or bug reports are appreciated as are example GPML files that do not convert properly. Finally, if you have any questions please feel free to drop a note in the comments below.

